We therefore examined the role of mitochondrial DNA mtDNA damage in human postmortem brain tissue and in in vivo and in vitro models of PD using a newly adapted histochemical assay for abasic sites and a quantitative polymerase chain reaction QPCR-based assay. Effect of Aβ42O on reactive oxygen species ROS production and DNA damage using mitoSOX staining and long-range PCR lesion assay respectively.
The mitochondrial genome mtGenome is multi-copy per cell the number of which varies greatly by cell type.
Mitochondrial dna damage assay. The quantitative polymerase chain reaction QPCR assay allows measurement of DNA damage in the mitochondrial and nuclear genomes without isolation of mitochondria. It also permits measurement of relative mitochondrial genome copy number. Finally it can be used for measurement of DNA repair in vivo when employed appropriately.
Given the crucial role of DNA damage in human health and disease it is important to be able to accurately measure both mitochondrial and nuclear DNA damage. This article describes a method based on a long-amplicon quantitative PCR-based assay that does not require a separate mitochondrial isolation step which can often be labor-intensive and generate artifacts. The detailed basic protocol.
There is a need for a rapid assay to identify agents that damage mitochondria because the mitochondrion may be an important target for numerous environmental mitotoxins. Certainly at least one chemotherapeutic regimen CHOP therapy that includes doxorubicin can induce cardiomyopathy through mitochondrial genotoxicity in cardiac muscle cells. Yeast cells 15 x 106-107 in water are spread.
DNA damage can cause and result from oxidative stress and mitochondrial impairment both of which are implicated in the pathogenesis of Parkinsons disease PD. We therefore examined the role of mitochondrial DNA mtDNA damage in human postmortem brain tissue and in in vivo and in vitro models of PD using a newly adapted histochemical assay for abasic sites and a quantitative polymerase chain reaction QPCR-based assay. We identified the molecular identity of mtDNA damage.
This assay has been extensively used to measure the integrity of both nuclear and mitochondrial genomes exposed to different genotoxins and has proved particularly valuable in identifying reactive oxygen species-mediated mitochondrial DNA mtDNA damage. QPCR can be used to quantify the formation of DNA damage as well as the kinetics of damage removal. One of the main strengths of the assay is that it permits monitoring the integrity of mtDNA.
This assay has been used extensively to measure the integrity of both nuclear and mitochondrial genomes exposed to different genotoxins and has proven to be particularly valuable in identifying reactive oxygen species-mediated mitochondrial DNA damage. QPCR can be used to quantify both the formation of DNA damage as well as the kinetics of damage removal. One of the main strengths of the assay is that it permits monitoring the integrity of mtDNA.
The smaller amplicon size greatly facilitates PCR optimization and allows greater flexibility in the use of detection dyes and a modified data analysis method simplifies the calculation of lesion frequency. The method was used to measure DNA damage in the nuclear and mitochondrial genomes of different tissues in zebrafish of different ages. We find that nuclear DNA damage generally increases with age and that the amount of mitochondrial DNA damage.
The mitochondrial genome mtGenome is multi-copy per cell the number of which varies greatly by cell type. Mitochondrial DNA mtDNA is highly susceptible to oxidative damage. Effect of Aβ42O on reactive oxygen species ROS production and DNA damage using mitoSOX staining and long-range PCR lesion assay respectively.
MtDNA repair activity was measured by non-homologous end joining NHEJ in vitro assay using mitochondria isolates and the expression and localization of NHEJ components were. Mitochondria repaired 50 of the damage after 1 h and by 6 h all the damage was repaired. Higher doses of GO-generated H202 or more extended treatment periods lead to mitochondrial DNA damage which was not repaired.
Mitochondrial function was monitored using the MTT 3 45-dimethylthiazol-2-yl25-diphenyltetrazolium bromide assay. The budding yeast Saccharomyces cerevisiae is a facile and informative model system in which to study such mtDNA oxidative damage because it is a unicellular eukaryotic facultative anaerobe that is conditionally dependent on mitochondrial oxidative phosphorylation for viability. Here we describe methods for quantifying oxidative mtDNA damage and mutagenesis in S.
Cerevisiae several of which. DNA damage in the mitochondrial genome was measured utilizing a PCR-based assay currently the most robust way of measuring damage in mtDNA 57. The quantitative polymerase chain reaction QPCR assay allows measurement of DNA damage in the mitochondrial and nuclear genomes without isolation of mitochondria.
It also permits measurement of relative mitochondrial genome copy number. Finally it can be used for measurement of DNA repair in vivo when employed appropriately. In this manuscript we briefly review the methodology of the QPCR.
In this study we found that curcumin induced DNA damage to both the mitochondrial and nuclear genomes in human hepatoma G2 cells. Using quantitative polymerase chain reaction and immunocytochemistry staining of 8-hydroxydeoxyguanosine we demonstrated that curcumin induced dose-dependent damage in both the mitochondrial and nuclear genomes and that the mitochondrial. The damage of mtDNA and nuclear DNA was analyzed by a long-range qPCR DNA lesion assay 24 h after treatment.
The PCR amplification of a large mtDNA segment 89 kb a small mtDNA segment 221 bp and the gene of polymerase β 122 kb was performed. DNA damage was documented as increased levels of γH2AX pCHK2 by Comet assay AIM2 induction and by increased DNA repair non-homologous end joining signaling. The DNA damage response DDR was not related to increased ROS levels.
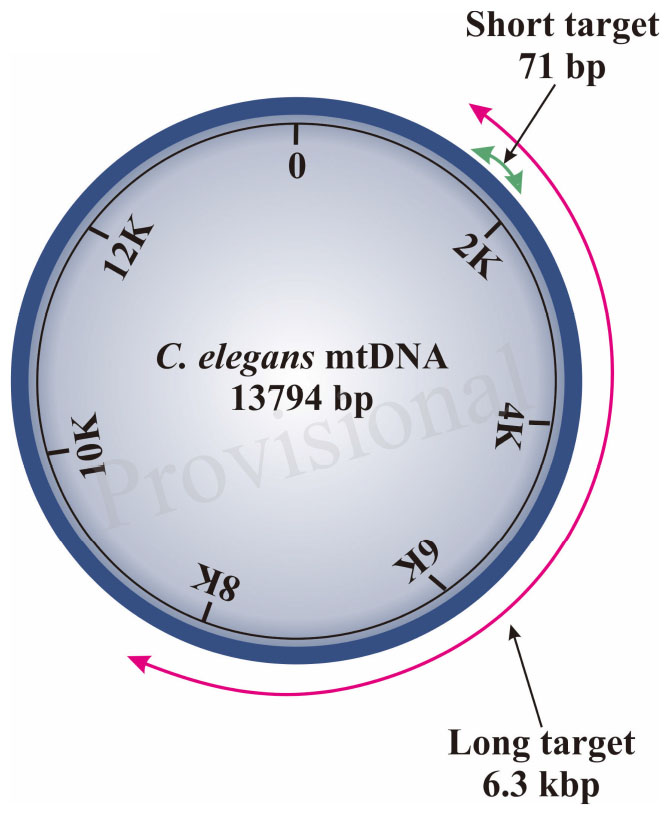
Schematic diagram of region amplified by short and long target using the S-XL-qRT-PCR DNA damage assay within C. The short target 71 bp is represented by a green arrow and the long amplicon 6300 bp is represented by the red arrow. Mitochondrial DNA is the small circular chromosome found inside mitochondria.
These organelles found in cells have often been called the powerhouse of the cell. The mitochondria and thus mitochondrial DNA are passed almost exclusively from mother to offspring through the egg cell. Electron microscopy reveals mitochondrial DNA in discrete foci.